Synthetic Data Generation for Pediatric Diabetes Research Using GANs and WGANs
DOI:
https://doi.org/10.17488/RMIB.46.1.3Keywords:
Generative Adversarial Networks, pediatric diabetes, synthetic data generation, Wasserstein GANsAbstract
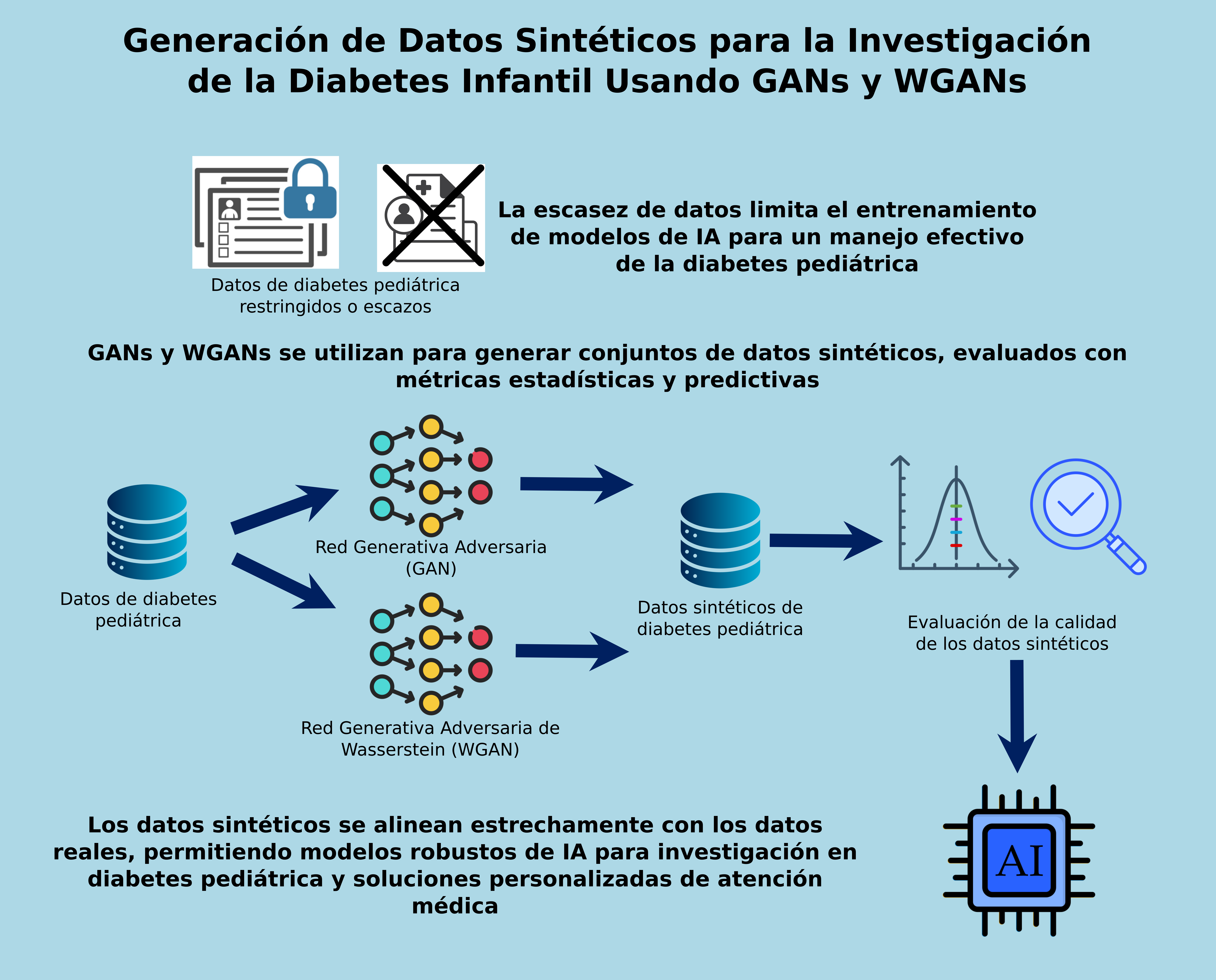
Pediatric diabetes research is often constrained by data scarcity, hindering the development of accurate predictive models for clinical applications. This study addresses this limitation by evaluating the effectiveness of Generative Adversarial Networks (GANs) and Wasserstein GANs (WGANs) in generating synthetic datasets that replicate the statistical properties of real pediatric diabetes data. A structured methodology was applied, incorporating preprocessing, model design, and dual evaluation metrics: Jensen-Shannon and Kullback-Leibler divergences for statistical fidelity, and a classification model to assess practical utility. Results demonstrate that both models produce high-fidelity synthetic datasets, with WGANs showing superior performance in capturing complex patterns due to improved training stability. Nonetheless, challenges remain in replicating the inherent variability of pediatric data, influenced by growth and developmental factors. This work highlights the potential of synthetic data to augment pediatric diabetes datasets, facilitating the development of robust and generalizable predictive models. Limitations include the dependency on initial data quality and the specificity of the models to pediatric datasets. By addressing critical gaps in data availability, this study contributes to advancing AI-driven healthcare solutions in pediatric diabetes research.
Downloads
References
D. J. G. Carrasco Ramírez, M. Islam, and I. H. Even, “Machine Learning Applications in Healthcare: Current Trends and Future Prospects,” J. Artif. Intell. Gen. Sci., vol. 1, no. 1, Art. no. 1, 2024, doi: https://doi.org/10.60087/jaigs.v1i1.33
A. Qayyum, J. Qadir, M. Bilal, and A. Al-Fuqaha, “Secure and Robust Machine Learning for Healthcare: A Survey,” IEEE Rev. Biomed. Eng., vol. 14, pp. 156–180, 2021, doi: https://doi.org/10.1109/RBME.2020.3013489
M. Javaid et al., “Significance of machine learning in healthcare: Features, pillars and applications,” Int. J. Intell. Netw., vol. 3, pp. 58–73, 2022, doi: https://doi.org/10.1016/j.ijin.2022.05.002
A. García-Domínguez et al., “Optimizing Clinical Diabetes Diagnosis through Generative Adversarial Networks: Evaluation and Validation,” Diseases, vol. 11, no. 4, 2023, art. no. 134, doi: https://doi.org/10.3390/diseases11040134
A. García-Domínguez et al., “Diabetes Detection Models in Mexican Patients by Combining Machine Learning Algorithms and Feature Selection Techniques for Clinical and Paraclinical Attributes: A Comparative Evaluation,” J. Diabetes Res., vol. 2023, 2023, art. no. 9713905, doi: https://doi.org/10.1155/2023/9713905
J. G. Elmore and C. I. Lee, “Data Quality, Data Sharing, and Moving Artificial Intelligence Forward,” JAMA Netw. Open, vol. 4, no. 8, 2021, art. no. e2119345, doi: https://doi.org/10.1001/jamanetworkopen.2021.19345
H. Sun et al., “IDF Diabetes Atlas: Global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045,” Diabetes Res. Clin. Pract., vol. 183, 2022, art. no. 109119, doi: https://doi.org/10.1016/j.diabres.2021.109119
C. W. Tsao et al., “Heart Disease and Stroke Statistics—2022 Update: A Report From the American Heart Association,” Circulation, vol. 145, no. 8, pp. e153–e639, 2022, doi: https://doi.org/10.1161/cir.0000000000001052
L. C. Stene and J. Tuomilehto, “Epidemiology of Type 1 Diabetes,” in Textbook of Diabetes, R.I.G. Holt and A. Flyvbjerg (Eds.)., Pondicherry, India: Wiley, 2024, ch. 4, pp. 41–54, doi: https://doi.org/10.1002/9781119697473.ch4
G. Imperatore et al., “Projections of Type 1 and Type 2 Diabetes Burden in the U.S. Population Aged <20 Years Through 2050: Dynamic modeling of incidence, mortality, and population growth,” Diabetes Care, vol. 35, no. 12, pp. 2515–2520, 2012, doi: https://doi.org/10.2337/dc12-0669
P. Bjornstad et al., “Youth-onset type 2 diabetes mellitus: an urgent challenge,” Nat. Rev. Nephrol., vol. 19, no. 3, pp. 168-184, 2023, doi: https://doi.org/10.1038/s41581-022-00645-1
J. Jordon et al., “Synthetic Data -- what, why and how?,” 2022, arXiv:2205.03257, doi: https://doi.org/10.48550/arXiv.2205.03257
V. C. Pezoulas et al., “Synthetic data generation methods in healthcare: A review on open-source tools and methods,” Comput. Struct. Biotechnol. J., vol. 23, pp. 2892–2910, 2024, doi: https://doi.org/10.1016/j.csbj.2024.07.005
I. Goodfellow et al., “Generative Adversarial Networks,” 2014, arXiv:1406.2661, doi: https://doi.org/10.48550/arXiv.1406.2661
M. Arjovsky, S. Chintala, L. Bottou, “Wasserstein Generative Adversarial Networks,” in Proceedings of the 34th International Conference on Machine Learning, Sydney, Australia, 2017, pp. 214-223, doi: https://doi.org/10.5555/3305381.3305404
M. L. Menéndez, J. A. Pardo, L. Pardo, and M. C. Pardo, “The Jensen-Shannon divergence,” J. Frankl. Inst., vol. 334, no. 2, pp. 307–318, 1997, doi: https://doi.org/10.1016/S0016-0032(96)00063-4
S. Kullback and R. A. Leibler, “On Information and Sufficiency,” Ann. Math. Stat., vol. 22, no. 1, pp. 79–86, 1951, doi: https://doi.org/10.1214/aoms/1177729694
P. A. Apellániz et al., “Synthetic Tabular Data Validation: A Divergence-Based Approach,” IEEE Access, vol. 12, pp. 103895–103907, 2024, doi: https://doi.org/10.1109/ACCESS.2024.3434582
X. Donath et al., “Next-generation sequencing identifies monogenic diabetes in 16% of patients with late adolescence/adult-onset diabetes selected on a clinical basis: a cross-sectional analysis,” BMC Med., vol. 17, no. 1, 2019, art. no. 132, doi: https://doi.org/10.1186/s12916-019-1363-0
Y. Park, D. Heider, and A.-C. Hauschild, “Integrative Analysis of Next-Generation Sequencing for Next-Generation Cancer Research toward Artificial Intelligence,” Cancers, vol. 13, no. 13, 2021, art. no. 3148, doi: https://doi.org/10.3390/cancers13133148
P. S. Paladugu et al., “Generative Adversarial Networks in Medicine: Important Considerations for this Emerging Innovation in Artificial Intelligence,” Ann. Biomed. Eng., vol. 51, pp. 2130-2142, 2023, doi: https://doi.org/10.1007/s10439-023-03304-z
A. Aggarwal, M. Mittal, and G. Battineni, “Generative adversarial network: An overview of theory and applications,” Int. J. Inf. Manag. Data Insights, vol. 1, no. 1, 2021, art no. 100004, doi: https://doi.org/10.1016/j.jjimei.2020.100004
C. O. S. Sorzano, J. Vargas, and A. Pascual, “A survey of dimensionality reduction techniques”, 2014, arXiv:1403.2877, doi: https://doi.org/10.48550/arXiv.1403.2877
G. T. Reddy et al., “Analysis of Dimensionality Reduction Techniques on Big Data,” IEEE Access, vol. 8, pp. 54776–54788, 2020, doi: https://doi.org/10.1109/ACCESS.2020.2980942
P. C. Austin, I. R. White, D. S. Lee, and S. van Buuren, “Missing Data in Clinical Research: A Tutorial on Multiple Imputation,” Can. J. Cardiol., vol. 37, no. 9, pp. 1322–1331, 2021, doi: https://doi.org/10.1016/j.cjca.2020.11.010
T. Emmanuel et al., “A survey on missing data in machine learning,” J. Big Data, vol. 8, no. 1, 2021, art. no. 140, doi: https://doi.org/10.1186/s40537-021-00516-9
R. S. Somasundaram and R. Nedunchezhian, “Evaluation of Three Simple Imputation Methods for Enhancing Preprocessing of Data with Missing Values,” Int. J. Comput. Appl., vol. 21, no. 10, pp. 14–19, 2011, doi: https://doi.org/10.5120/2619-3544
R. J. A. Little and D. B. Rubin, Statistical Analysis with Missing Data, 3rd ed., Hoboken, NJ, USA: Wiley, 2019, doi: https://doi.org/10.1002/9781119482260
C. K. Enders, Applied Missing Data Analysis, 2nd ed., Guilford Publications, 2022.
S. van Buuren, Flexible Imputation of Missing Data, 2nd ed., NY, USA: CRC Press, 2018, doi: https://doi.org/10.1201/9780429492259
A. Géron, Hands-On Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems, 2nd ed., O’Reilly Media, Inc., 2019.
S. G. K. Patro and K. K. Sahu, “Normalization: A Preprocessing Stage,” Int. Adv. Res. J. Sci. Eng. Technol., pp. 20–22, 2015, doi: https://doi.org/10.17148/IARJSET.2015.2305
D. Singh and B. Singh, “Investigating the impact of data normalization on classification performance,” Appl. Soft Comput., vol. 97, 2020, art. no. 105524, doi: https://doi.org/10.1016/j.asoc.2019.105524
L. Rahmad Ramadhan and Y. Anne Mudya, “A Comparative Study of Z-Score and Min-Max Normalization for Rainfall Classification in Pekanbaru,” J. Data Sci., vol. 2024, 2024, art. no. 4. [Online]. Available: https://iuojs.intimal.edu.my/index.php/jods/article/view/446
M. Arjovsky, S. Chintala, and L. Bottou, “Wasserstein GAN,” 2017, arXiv:7101.07875, doi: https://doi.org/10.48550/arXiv.1701.07875
I. Goodfellow et al., “Generative Adversarial Nets,” in Advances in Neural Information Processing Systems, 2014. [Online]. Available: https://proceedings.neurips.cc/paper_files/paper/2014/file/5ca3e9b122f61f8f06494c97b1afccf3-Paper.pdf
T. Salimans et al., “Improved Techniques for Training GANs,” in Advances in Neural Information Processing Systems, 2016. [Online]. Available: https://proceedings.neurips.cc/paper_files/paper/2016/file/8a3363abe792db2d8761d6403605aeb7-Paper.pdf
A. Radford, L. Metz, and S. Chintala, “Unsupervised Representation Learning with Deep Convolutional Generative Adversarial Networks,” 2016, arXiv:1511.06434, doi: https://doi.org/10.48550/arXiv.1511.06434
S. Jenni and P. Favaro, “On Stabilizing Generative Adversarial Training With Noise,” in 2019 IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 2019, pp. 12137–12145, doi: https://doi.org/10.1109/CVPR.2019.01242
X. Glorot and Y. Bengio, “Understanding the difficulty of training deep feedforward neural networks”, in Proceedings of the Thirteenth International Conference on Artificial Intelligence and Statistics, Sardinia, Italy, 2010. [Online]. Available: https://proceedings.mlr.press/v9/glorot10a/glorot10a.pdf
I. Gulrajani et al., “Improved Training of Wasserstein GANs,” in NIPS'17: Proceedings of the 31st International Conference on Neural Information Processing Systems, Long Beach, California, USA, 2017. [Online]. Available: https://dl.acm.org/doi/10.5555/3295222.3295327
I. Goodfellow, Y. Bengio, and A. Courville, Deep Learning. MIT Press, 2016.
J. Lin, “Divergence measures based on the Shannon entropy,” IEEE Trans. Inf. Theory, vol. 37, no. 1, pp. 145–151, 1991, doi: https://doi.org/10.1109/18.61115
M. Lucic et al., “Are GANs Created Equal? A Large-Scale Study,” in NIPS'18: Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montreal, Canada, 2018. [Online]. Available: https://dl.acm.org/doi/10.5555/3326943.3327008
A. Borji, “Pros and cons of GAN evaluation measures,” Comput. Vis. Image Underst., vol. 179, pp. 41–65, 2019, doi: https://doi.org/10.1016/j.cviu.2018.10.009
S. Arora et al., “Generalization and Equilibrium in Generative Adversarial Nets (GANs),” in ICML'17: Proceedings of the 34th International Conference on Machine Learning - Volume 70, Sydney, Australia, 2017, pp. 224–232. [Online]. Available: https://dl.acm.org/doi/10.5555/3305381.3305405
M. S. M. Sajjadi et al., “Assessing Generative Models via Precision and Recall,” in NIPS'18: Proceedings of the 32nd International Conference on Neural Information Processing Systems, Montreal, Canada, 2018. [Online]. Available: https://dl.acm.org/doi/10.5555/3327345.3327429
S. Nowozin, B. Cseke, and R. Tomioka, “f-GAN: Training Generative Neural Samplers using Variational Divergence Minimization,” in NIPS'16: Proceedings of the 30th International Conference on Neural Information Processing Systems, Barcelona, España, 2016. [Online]. Available: https://dl.acm.org/doi/10.5555/3157096.3157127
L. Breiman, “Random Forests,” Mach. Learn., vol. 45, no. 1, pp. 5–32, 2001, doi: https://doi.org/10.1023/A:1010933404324
A. Liaw and M. Wiener, “Classification and Regression by randomForest,” R News, vol. 2, pp. 18-22, 2002. [Online]. Available: https://www.r-project.org/doc/Rnews/Rnews_2002-3.pdf
T. K. Ho, “Random decision forests,” in Proceedings of 3rd International Conference on Document Analysis and Recognition, Montreal, QC, Canada, 1995, pp. 278-282, doi: https://doi.org/10.1109/ICDAR.1995.598994
F. Pedregosa et al., “Scikit-learn: Machine Learning in Python,” J. Mach. Learn. Res., vol. 12, pp. 2825-2830, 2011. [Online]. Available: https://dl.acm.org/doi/10.5555/1953048.2078195
C. Shorten and T. M. Khoshgoftaar, “A survey on Image Data Augmentation for Deep Learning,” J. Big Data, vol. 6, no. 1, 2019, art. no. 60, doi: https://doi.org/10.1186/s40537-019-0197-0
E. Choi et al., “Generating Multi-label Discrete Patient Records using Generative Adversarial Networks,” 2018, arXiv:1703.06490, doi: https://doi.org/10.48550/arXiv.1703.06490
A. Goncalves et al., “Generation and evaluation of synthetic patient data,” BMC Med. Res. Methodol., vol. 20, no. 1, 2020, art. no. 108, doi: https://doi.org/10.1186/s12874-020-00977-1
L. Xu, et al., “Modeling Tabular data using Conditional GAN,” 2019, arXiv:1907.00503, doi: https://doi.org/10.48550/arXiv.1907.00503
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2025 Revista Mexicana de Ingenieria Biomedica

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Upon acceptance of an article in the RMIB, corresponding authors will be asked to fulfill and sign the copyright and the journal publishing agreement, which will allow the RMIB authorization to publish this document in any media without limitations and without any cost. Authors may reuse parts of the paper in other documents and reproduce part or all of it for their personal use as long as a bibliographic reference is made to the RMIB. However written permission of the Publisher is required for resale or distribution outside the corresponding author institution and for all other derivative works, including compilations and translations.







