UMInSe: Método no Supervisado para la Segmentación y Detección de Instrumentos Quirúrgicos Basado en K-means
DOI:
https://doi.org/10.17488/RMIB.45.3.2Palabras clave:
base de datos JIGSAWS, K-means, segmentación instrumentos quirúrgicos, segmentación no supervisadaResumen
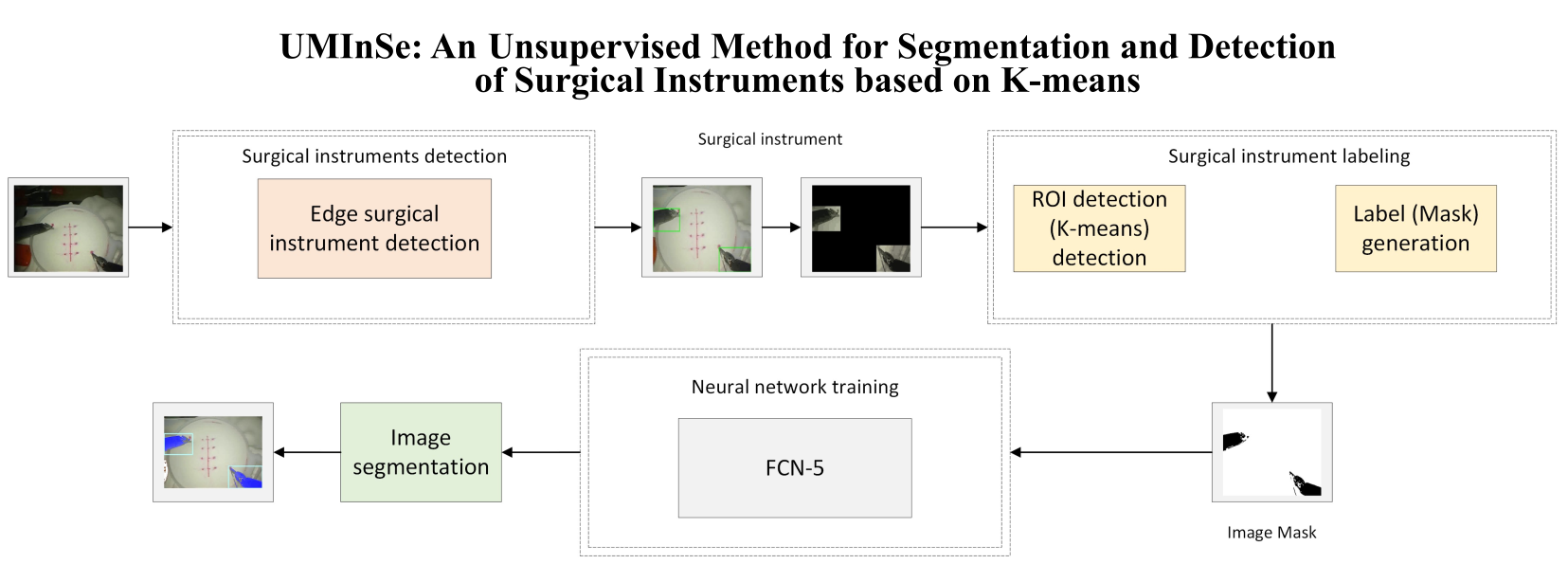
La segmentación de instrumentos quirúrgicos en imágenes es crucial para mejorar la precisión y eficiencia en cirugía, pero actualmente depende de anotaciones manuales costosas y laboriosas. Un enfoque no supervisado es una solución prometedora para este desafío. Este artículo introduce un método de segmentación de instrumentos quirúrgicos utilizando aprendizaje automático no supervisado, basado en el algoritmo K-means, para identificar Regiones de Interés (ROI) en imágenes y crear el ground truth de las imágenes para el entrenamiento de redes neuronales. La corrección Gamma ajusta el brillo de la imagen y mejora la identificación de áreas que contienen instrumentos quirúrgicos. El algoritmo K-means agrupa píxeles similares y detecta las ROI a pesar de los cambios en la iluminación, logrando una segmentación eficiente a pesar de las variaciones en la iluminación de la imagen y los objetos obstructores. Por lo tanto, la red neuronal generaliza el aprendizaje de las características de la imagen para la segmentación de instrumentos en diferentes tareas. Los resultados experimentales utilizando las bases de datos JIGSAWS y EndoVis demuestran la efectividad y robustez del método, con un error mínimo (0.0297) y alta precisión (0.9602). Estos resultados subrayan la precisión en la detección y segmentación de instrumentos quirúrgicos, lo cual es crucial para automatizar la detección de instrumentos en procedimientos quirúrgicos sin conjuntos de datos pre-etiquetados. Además, esta técnica podría aplicarse en aplicaciones quirúrgicas como la evaluación de habilidades del cirujano y la planificación de movimientos de robots, donde la detección precisa de instrumentos es indispensable.
Descargas
Citas
X. Wang, L. Wang, X. Zhong, C. Bai, X. Huang, R. Zhao, and M. Xia, “Pal-Net: A modified U-Net of reducing semantic gap for surgical instrument segmentation,” IET Image Process., vol. 15, no. 12, pp. 2959-2969, 2021, doi: https://doi.org/10.1049/ipr2.12283
S. Nema and L. Vachhani, “Unpaired deep adversarial learning for multi-class segmentation of instruments in robot-assisted surgical videos,” Int. J. Med. Robot., vol. 19, no. 4, 2023, no. art. e2514, doi: https://doi.org/10.1002/rcs.2514
E.-J. Lee, W. Plishker, X. Liu, S. S. Bhattacharyya, and R. Shekhar, “Weakly supervised segmentation for real-time surgical tool tracking,” Healthc. Technol. Lett., vol. 6, no. 6, pp. 231-236, 2019, doi: https://doi.org/10.1049/htl.2019.0083
N. Ahmidi, L. Tao, S. Sefati, Y. Gao, et al., “A Dataset and Benchmarks for Segmentation and Recognition of Gestures in Robotic Surgery, IEEE Trans. Biomed. Eng., vol. 64, no. 9, pp. 2025-2041, 2017, doi: https://doi.org/10.1109/TBME.2016.2647680
F. Chadebecq, F. Vasconcelos, E. Mazomenos and D. Stoyanov, “Computer Vision in the Surgical Operating Room,” Visc. Med., vol. 36, no. 6, pp. 456-462, 2020, doi: https://doi.org/10.1159/000511934
D. Psychogyios, E. Mazomenos, F. Vasconcelos, and D. Stoyanov, “MSDESIS: Multitask Stereo Disparity Estimation and Surgical Instrument Segmentation,” IEEE Trans. Med. Imaging, vol. 41, no. 11, pp. 3218-3230, 2022 doi: https://doi.org/10.1109/tmi.2022.3181229
C. González, L. Bravo-Sanchéz, and P. Arbeláez, “Surgical instrument grounding for robot-assisted interventions,” Comput. Methods Biomech. Biomed. Eng.: Imaging Vis., vol. 10, no. 3, pp. 299-307, 2022, doi: https://doi.org/10.1080/21681163.2021.2002725
W. Burton, C. Myers, M. Rutherford, and P. Rullkoetter, “Evaluation of single-stage vision models for pose estimation of surgical instruments,” Int. J. Comput. Assist. Radiol. Surg., vol. 18, no. 12, pp. 2125-2142, 2023, doi: https://doi.org/10.1007/s11548-023-02890-6
Y. Jin, Y. Yu, C. Chen, Z. Zhao, P.-A. Heng, and D. Stoyanov, “Exploring Intra- and Inter-Video Relation for Surgical Semantic Scene Segmentation,” IEEE Tran. Med. Imaging, vol. 41, no. 11, pp. 2991-3002, 2022, doi: https://doi.org/10.1109/TMI.2022.3177077
M. Attia, M. Hossny, S. Nahavandi, and H. Asadi, “Surgical tool segmentation using a hybrid deep CNN-RNN auto encoder-decoder,” 2017 IEEE International Conference on Systems, Man, and Cybernetics (SMC), Banff, AB, Canada, 2017, pp. 3373-3378, doi: https://doi.org/10.1109/SMC.2017.8123151
D. Papp, R. N. Elek, and T. Haidegger, “Surgical Tool Segmentation on the JIGSAWS Dataset for Autonomous Image-based Skill Assessment,” 2022 IEEE 10th Jubilee International Conference on Computational Cybernetics and Cyber-Medical Systems (ICCC), Reykjavík, Iceland, 2022, doi: https://doi.org/10.1109/ICCC202255925.2022.9922713
M. Daneshgar Rahbar and S. Z. Mousavi Mojab, “Enhanced U-Net with GridMask (EUGNet): A Novel Approach for Robotic Surgical Tool Segmentation,” J. Imaging, vol. 9, no. 12, 2023, no art. 282, doi: https://doi.org/10.3390/jimaging9120282
E. Colleoni, D. Psychogyios, B. Van Amsterdam, F. Vasconcelos, and D. Stoyanov, “SSIS-Seg: Simulation-Supervised Image Synthesis for Surgical Instrument Segmentation,” IEEE Trans. Med. Imaging, vol. 41, no. 11, pp. 3074-3086, 2022, doi: https://doi.org/10.1109/tmi.2022.3178549
P. Deepika, K. Udupa, M. Beniwal, A. M. Uppar, V. Vikas, and M. Rao, “Automated Microsurgical Tool Segmentation and Characterization in Intra-Operative Neurosurgical Videos,” 2022 44th Annual International Conference of the IEEE Engineering in Medicine & Biology Society, Glasgow, Scotland, United Kingdom, 2022, pp. 2110-2114, doi: https://doi.org/10.1109/EMBC48229.2022.9871838
G. Leifman, A. Aides, T. Golany, D. Freedman, and E. Rivlin, “Pixel-accurate Segmentation of Surgical Tools based on Bounding Box Annotations,” 2022 26th International Conference on Pattern Recognition (ICPR), Montreal, QC, Canada, 2022, pp. 5096-5103, doi: https://doi.org/10.1109/ICPR56361.2022.9956530
R. Mishra, A. Thangamani, K. Palle, P. V. Prasad, B. Mallala, and T.R. V. Lakshmi, “Adversarial Transfer Learning for Surgical Instrument Segmentation in Endoscopic Images,” 2023 IEEE International Conference on Paradigm Shift in Information Technologies with Innovative Applications in Global Scenario (ICPSITIAGS), Indore, India, 2023, pp. 28-34, doi: https://doi.org/10.1109/ICPSITIAGS59213.2023.10527520
A. Lou, K. Tawfik, X. Yao, Z. Liu, and J. Noble, “Min-Max Similarity: A Contrastive Semi-Supervised Deep Learning Network for Surgical Tools Segmentation,” IEEE Trans. Med. Imaging, vol 42, no. 10, pp. 2023, doi: https://doi.org/10.1109/TMI.2023.3266137
E. Colleoni and D. Stoyanov, “Robotic Instrument Segmentation With Image-to-Image Translation,” IEEE Robot. Autom. Lett., vol. 6, no. 2, pp. 935-942, 2021, doi: https://doi.org/10.1109/LRA.2021.3056354
D. Jha, S. Ali, N. K. Tomar, M. A. Riegler, and D. Johansen, “Exploring Deep Learning Methods for Real-Time Surgical Instrument Segmentation in Laparoscopy,” 2021 IEEE EMBS International Conference on Biomedical and Health Informatics (BHI), Athens, Greece, 2021, doi: https://doi.org/10.1109/BHI50953.2021.9508610
M. Allan, S. Ourselin, D. J. Hawkes, J. D. Kelly and D. Stoyanov, “3-D Pose Estimation of Articulated Instruments in Robotic Minimally Invasive Surgery,” IEEE Trans. Med. Imaging, vol. 37, no. 5, pp. 1204-1213, pp. 1204-1213, 2018, doi: https://doi.org/10.1109/tmi.2018.2794439
L. Wang, C. Zhou, Y. Cao, R. Zhao, and K. Xu, “Vision-Based Markerless Tracking for Continuum Surgical Instruments in Robot-Assisted Minimally Invasive Surgery,” IEEE Robot. Autom. Lett., vol. 8, no. 11, pp. 7202-7209, 2023, doi: https://doi.org/10.1109/LRA.2023.3315229
L. Yu, P. Wang, X. Yu, Y. Yan, and Y. Xia, “A Holistically-Nested U-Net: Surgical Instrument Segmentation Based,” J. Digit. Imaging, vol. 33, no. 2, pp. 341-347, 2020, doi: https://doi.org/10.1007%2Fs10278-019-00277-1
M. Xue and L. Gu, “Surgical instrument segmentation method based on improved MobileNetV2 network,” 2021 6th International Symposium on Computer and Information Processing Technology (ISCIPT), Changsha, China, 2021, pp. 744-747, doi: https://doi.org/10.1109/ISCIPT53667.2021.00157
B. Baby, D. Thapar, M. Chasmai, T. Banerjee, et al., “From Forks to Forceps: A New Framework for Instance Segmentation of Surgical Instruments,” 2023 IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Waikoloa, HI, USA, 2023, doi: https://doi.org/10.1109/WACV56688.2023.00613
T. Streckert, D. Fromme, M. Kaupenjohann and J. Thiem, “Using Synthetic Data to Increase the Generalization of a CNN for Surgical Instrument Segmentation,” 2023 IEEE 12th International Conference on Intelligent Data Acquisition and Advanced Computing Systems: Technology and Applications (IDAACS), Dortmund, Germany, 2023, pp. 336-340, doi: https://doi.org/10.1109/IDAACS58523.2023.10348781
Y. Yamada, J. Colan, A. Davila, and Yasuhisa Hasegawa, “Task Segmentation Based on Transition State Clustering for Surgical Robot Assistance,” 2023 8th International Conference on Control and Robotics Engineering (ICCRE), Niigata, Japan, 2023, pp. 260-264, doi: https://doi.org/10.1109/ICCRE57112.2023.10155581
Z. Zhang, B. Rosa, and F. Nageotte, “Surgical Tool Segmentation Using Generative Adversarial Networks With Unpaired Training Data,” IEEE Robot. Autom. Lett., vol 6, no. 4, pp. 6266-6273, 2021, doi: https://doi.org/10.1109/LRA.2021.3092302
A. Qayyum, M. Bilal, J. Qadir, M. Caputo, et al., “SegCrop: Segmentation-based Dynamic Cropping of Endoscopic Videos to Address Label Leakage in Surgical Tool Detection,” 2023 IEEE 20th International Symposium on Biomedical Imaging (ISBI), Cartagena, Colombia, 2023, doi: https://doi.org/10.1109/ISBI53787.2023.10230822
Y. Gao, S. Vedula, C. E. Reiley, N. Ahmidi, et al., “JHU-ISI Gesture and Skill Assessment Working Set (JIGSAWS): A Surgical Activity Dataset for Human Motion Modeling,” Modeling and Monitoring of Computer Assisted Interventions (M2CAI), Boston, United State, 2014.
L. Maier-Hein, S. Mersmann, D. Kondermann, S. Bodenstedt, et al., “Can Masses of Non-Experts Train Highly Accurate Image Classifiers? A crowdsourcing approach to instrument segmentation in laparoscopic images,” Med. Image Comput. Comput. Assist. Interv., vol. 17, pp. 438-445, 2014, doi: https://doi.org/10.1007/978-3-319-10470-6_55
A. Murali, D. Alapatt, P. Mascagni, A. Vardazaryan, et al., “The Endoscapes Dataset for Surgical Scene Segmentation, Object Detection, and Critical View of Safety Assessment: Official Splits and Benchmark,” 2023, arXiv:2312.12429, doi: https://doi.org/10.48550/arXiv.2312.12429
M. Ju, D. Zhang, and Y. J. Guo, “Gamma-Correction-Based Visibility Restoration for Single Hazy Images,” IEEE Signal Process. Lett., vol. 25, no. 7, pp. 1084–1088, 2018, doi: https://doi.org/10.1109/LSP.2018.2839580
R. Silpasai, H. Singh, A. Kumarl and L. Balyan, “Homomorphically Rectified Tile-wise Equalized Adaptive Gamma Correction for Histopathological Color Image Enhancement,” 2018 Conference on Information and Communication Technology (CICT), Jabalpur, India, 2018, doi: https://doi.org/10.1109/INFOCOMTECH.2018.8722364
H. Ye, S. Yan and P. Huang, “2D Otsu image segmentation based on cellular genetic algorithm,” 9th International Conference on Communication Software and Networks (ICCSN), Guangzhou, China, 2017, pp. 1313-1316, doi: https://doi.org/10.1109/ICCSN.2017.8230322
P. Flach, Machine Learning: The Art and Science of Algorithms that Make Sense of Data, 1st Ed. Edinburgh, United Kingdom: Cambridge University Press, 2012.
A. C. Müller and S. Guido, Introduction to Machine Learning with Python, United States of America: O´Reilly, 2017.
P. Yin, R. Yuan, Y. Cheng and Q. Wu, “Deep Guidance Network for Biomedical,” IEEE Access, vol. 8, pp. 116106-116116, 2020, doi: https://doi.org/10.1109/ACCESS.2020.3002835
I. El rube’, “Image Color Reduction Using Progressive Histogram Quantization and K-means Clustering,” 2019 International Conference on Mechatronics, Remote Sensing, Information Systems and Industrial Information Technologies (ICMRSISIIT), Ghana, 2020, doi: https://doi.org/10.1109/ICMRSISIIT46373.2020.9405957
R. E. Arevalo-Ancona and M. Cedillo-Hernandez, “Zero-Watermarking for Medical Images Based on Regions of Interest Detection using K-Means Clustering and Discrete Fourier Transform,” Int. J. Adv. Comput. Sci. Appl., vol. 14, no. 6, 2023, doi: https://dx.doi.org/10.14569/IJACSA.2023.0140662
P. Sharma, “Advanced Image Segmentation Technique using Improved K Means Clustering Algorithm with Pixel Potential,” 2020 Sixth International Conference on Parallel, Distributed and Grid Computing (PDGC), Waknaghat, India, 2020, pp. 561-565, doi: https://doi.org/10.1109/PDGC50313.2020.9315743
Descargas
Publicado
Cómo citar
Número
Sección
Licencia
Derechos de autor 2024 Revista Mexicana de Ingenieria Biomedica

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial 4.0.
Una vez que el artículo es aceptado para su publicación en la RMIB, se les solicitará al autor principal o de correspondencia que revisen y firman las cartas de cesión de derechos correspondientes para llevar a cabo la autorización para la publicación del artículo. En dicho documento se autoriza a la RMIB a publicar, en cualquier medio sin limitaciones y sin ningún costo. Los autores pueden reutilizar partes del artículo en otros documentos y reproducir parte o la totalidad para su uso personal siempre que se haga referencia bibliográfica al RMIB. No obstante, todo tipo de publicación fuera de las publicaciones académicas del autor correspondiente o para otro tipo de trabajos derivados y publicados necesitaran de un permiso escrito de la RMIB.











