Performance Evaluation of Biomedical Time Series Transformation Methods for Classification Tasks
DOI:
https://doi.org/10.17488/RMIB.44.4.7Keywords:
biomedical data, classification, convolutional neural networks, time series, transformationsAbstract
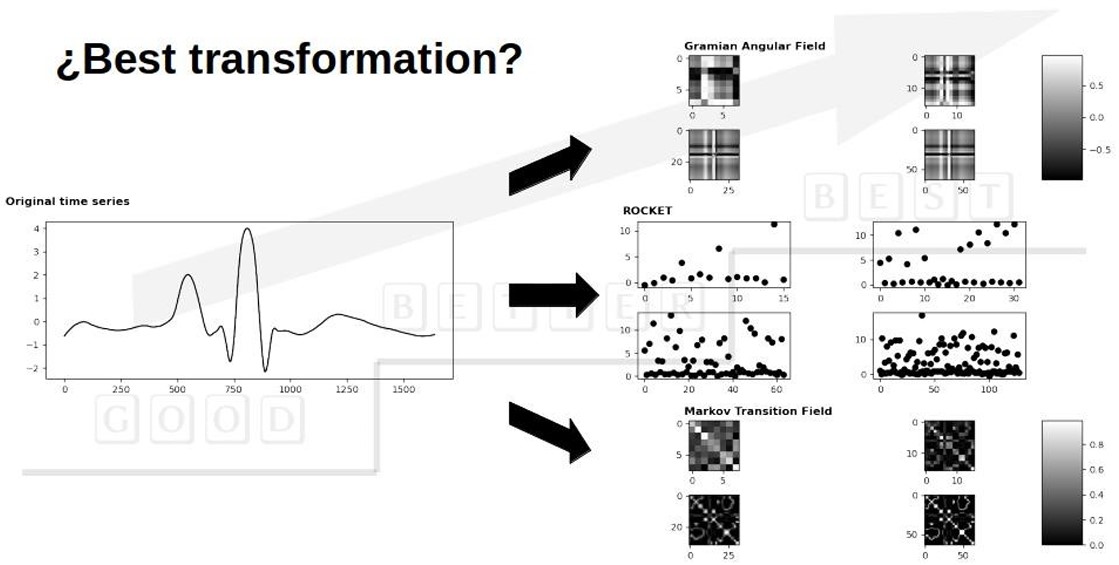
The extraction of time series features is essential across various fields, yet it remains a challenging endeavor. Therefore, it's crucial to identify appropriate methods capable of extracting pertinent information that can significantly enhance classification performance. Among these methods are those that translate time series into different domains. This study investigates three distinct time series transformation approaches for addressing time series classification challenges within biomedical data. The first method involves a response vector transformation, while the other two employ image transformation techniques: RandOm Convolutional KErnel Transform (ROCKET), Gramian Angular Fields, and Markov Transition Fields. These transformation methods were applied to five biomedical datasets, exploring various format configurations to ascertain the optimal representation technique and configuration for input, which in turn improves classification performance. Evaluations were conducted on the effectiveness of these methods in conjunction with two classification algorithms. The outcomes underscore the significance of these time series transformation techniques as facilitators for enhanced classification algorithms documented in current literature.
Downloads
References
M. Middlehurst, P. Schäfer, and A. Bagnall, “Bake off redux: a review and experimental evaluation of recent time series classification algorithms,” 2023, arXiv:2304.13029, doi: https://doi.org/10.48550/arXiv.2304.13029
H. Ismail Fawaz, G. Forestier, J. Weber, L. Idoumghar, and P.-A. Muller, “Deep learning for time series classification: a review,” Data Min. Knowl. Discov., vol. 33, no. 4, pp. 917–963, 2019, doi: https://doi.org/10.1007/s10618-019-00619-1
C. Li, J. Xiong, X. Zhu, Q. Zhang, and S. Wang, “Fault Diagnosis Method Based on Encoding Time Series and Convolutional Neural Network,” IEEE Access, vol. 8, pp. 165232–165246, 2020, doi: https://doi.org/10.1109/ACCESS.2020.3021007
G. R. Garcia, G. Michau, M. Ducoffe, J. Sen Gupta, and O. Fink, “Temporal signals to images: Monitoring the condition of industrial assets with deep learning image processing algorithms,” Proc. Inst. Mech. Eng. O J. Risk Reliab., vol. 236, no. 4, pp. 617–627, 2022, doi: https://doi.org/10.1177/1748006X21994446
J. Lines, S. Taylor, and A. Bagnall, “Hive-cote: The hierarchical vote collective of transformation-based ensembles for time series classification,” in 2016 IEEE 16th international conference on data mining (ICDM), Barcelona, Spain, 2016, pp. 1041–1046, doi: https://doi.org/10.1109/ICDM.2016.0133
Z. Wang, W. Yan, and T. Oates, “Time series classification from scratch with deep neural networks: A strong baseline,” in 2017 International Joint Conference on Neural Networks (IJCNN), Anchorage, AK, USA, 2017, pp. 1578–1585, doi: https://doi.org/10.1109/IJCNN.2017.7966039
H. Ismail Fawaz, B. Lucas, G. Forestier, C. Pelletier, et al., “Inceptiontime: Finding alexnet for time series classification,” Data Min. Knowl. Discov., vol. 34, no. 6, pp. 1936–1962, 2020, doi: https://doi.org/10.1007/s10618-020-00710-y
H. A. Dau, A. Bagnall, K. Kamgar, C.-C. M. Yeh, Y. Zhu, S. Gharghabi, C. A. Ratanamahatana, E. Keogh, “The UCR time series archive,” IEEE/CAA J. Autom. Sin., vol. 6, no. 6, pp. 1293–1305, 2019, doi: https://doi.org/10.1109/JAS.2019.1911747
A. Dempster, F. Petitjean, and G. I. Webb, “ROCKET: exceptionally fast and accurate time series classification using random convolutional kernels,” Data Min. Knowl. Discov., vol. 34, no. 5, pp. 1454–1495, 2020, doi: https://doi.org/10.1007/s10618-020-00701-z
Z. Wang and T. Oates, “Imaging time-series to improve classification and imputation,” 2015, arXiv:1506.00327, 2015, doi: https://doi.org/10.48550/arXiv.1506.00327
J. Faouzi and H. Janati, “pyts: A Python Package for Time Series Classification,” J. Mach. Learn. Res., vol. 21, no. 46, pp. 1–6, 2020. [Online]. Available: http://jmlr.org/papers/v21/19-763.html
F. Chollet, “Keras.” 2015, GitHub. [Online]. Available: https://github.com/fchollet/keras
F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, et al., “Scikit-learn: Machine learning in Python,” J. Mach. Learn. Res., vol. 12, no. 85, 2825-2830. [Online]. Available: http://www.jmlr.org/papers/v12/pedregosa11a.html
Tanu and D. Kakkar, “Accounting for order-frame length tradeoff of Savitzky-Golay smoothing filters,” in 2018 5th International Conference on Signal Processing and Integrated Networks (SPIN), Noida, India, 2018, pp. 805–810, doi: https://doi.org/10.1109/SPIN.2018.8474194
X. Li, Y. Kang, and F. Li, “Forecasting with time series imaging,” Expert. Syst. Appl., vol. 160, art. no. 113680, 2020, doi: https://doi.org/10.1016/j.eswa.2020.113680
A. M. Gonzalez-Zapata, L. G. de la Fraga, B. Ovilla-Martinez, E. Tlelo-Cuautle, and I. Cruz-Vega, “Enhanced FPGA implementation of Echo State Networks for chaotic time series prediction,” Integration, vol. 92, pp. 48–57, 2023, doi: https://doi.org/10.1016/j.vlsi.2023.05.002
A. Ben Said and A. Erradi, “Deep-Gap: A deep learning framework for forecasting crowdsourcing supply-demand gap based on imaging time series and residual learning,” 2019, arXiv:1911.07625, doi: https://doi.org/10.48550/arXiv.1911.07625
W. Ni, C. Zhang, T. Liu, Q. Zeng, L. Xu, and H. Wang, “An efficient astronomical seeing forecasting method by random convolutional Kernel transformation,” Eng. Appl. Artif. Intell., vol. 127, art. no. 107259, 2024, doi: https://doi.org/10.1016/j.engappai.2023.107259
J. Marco-Blanco and R. Cuevas, “Time Series Clustering With Random Convolutional Kernels,” 2023, arXiv:2305.10457, doi: https://doi.org/10.48550/arXiv.2305.10457
B. Dhariyal, T. Le Nguyen, and G. Ifrim, “Back to Basics: A Sanity Check on Modern Time Series Classification Algorithms,” 2023, arXiv:2308.07886, doi: https://doi.org/10.48550/arXiv.2308.07886
M. Middlehurst, J. Large, M. Flynn, J. Lines, A. Bostrom, and A. Bagnall, “HIVE-COTE 2.0: a new meta ensemble for time series classification,” Mach. Learn., vol. 110, no. 11–12, pp. 3211–3243, 2021, doi: https://doi.org/10.1007/s10994-021-06057-9
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Revista Mexicana de Ingenieria Biomedica

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Upon acceptance of an article in the RMIB, corresponding authors will be asked to fulfill and sign the copyright and the journal publishing agreement, which will allow the RMIB authorization to publish this document in any media without limitations and without any cost. Authors may reuse parts of the paper in other documents and reproduce part or all of it for their personal use as long as a bibliographic reference is made to the RMIB. However written permission of the Publisher is required for resale or distribution outside the corresponding author institution and for all other derivative works, including compilations and translations.







